Scientists smash thousands of proteins to find four ‘Legos of life’
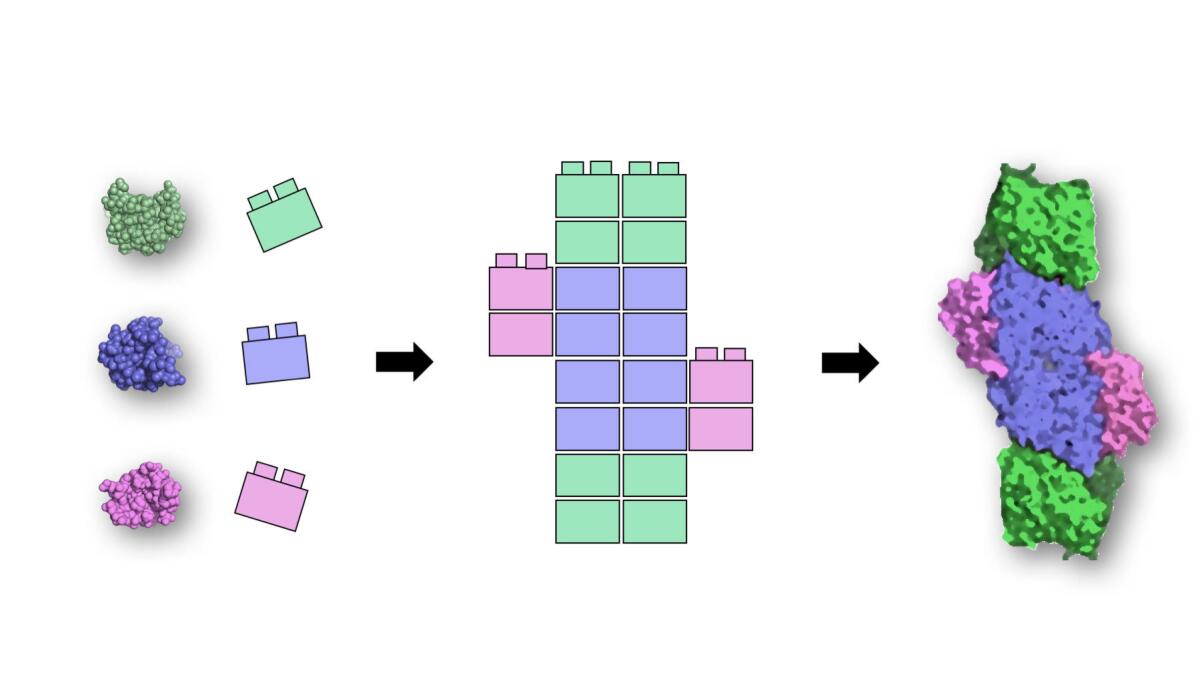
By “smashing” proteins and looking at the broken bits, scientists at Rutgers University say they’ve discovered four basic building blocks that can be stacked like Legos to build all kinds of different proteins.
The results described in Proceedings of the National Academy of Sciences could help researchers better understand the origins of life and design new biomolecules with special industrial and medical uses.
“We’ve developed methods for really looking at the deep-time evolution of proteins,” said senior author Vikas Nanda, a biochemist at Rutgers University.
Many scientists think that some of the earliest forms of life would have hung out where natural electric currents exist — at the bottom of the sea floor, for example, where hydrothermal vents spew material into the ocean.
Ancient microbes would have needed special proteins to take advantage of those energy sources. These metal-bearing “metalloproteins” would have been able to carry and move electrons around in specific ways. But what exactly did such proteins look like, some 4 billion years ago?
It’s hard to judge by what’s in modern-day microbes because their proteins are pretty complex, Nanda said. He pointed to proteins like nitrogenase, an essential enzyme that takes nitrogen and makes ammonia, which is then used to make DNA and amino acids.
“You couldn’t imagine that complex nanomachine just emerging out of the primordial soup and just coming into existence,” Nanda said. “There had to have been simpler intermediates. But the challenge is we don’t have any fossil record of what proteins look like. All we have is the modern proteins, and we have to somehow infer what the simpler proteins may have looked like.”
The average protein today is made up of around 200 to 250 amino acids, and still larger proteins can be made out of those amino acid chains. Nanda and his colleagues wanted to figure out which pieces within those chains truly did the work, and thus might resemble more ancient proteins from early life.
To get at this question, the researchers dug through the university’s RCSB Protein Data Bank to analyze the atomic 3-D structure of 9,500 proteins. Using computers, they picked through the amino acid chains, looking for common subunits shared across different proteins that seemed to have similar functions.
It was basically like trying to understand a radio’s most essential parts by taking a bunch of radios, smashing them with a hammer and then comparing the fragments, Nanda said.
“The question then is, what are the useful pieces and what’s just junk?” he said. “When we looked at the pieces, we’d find something that looks like a transistor or we would find something that looks like a tuning capacitor for the dial, and we would categorize them into these different classes.”
Sure enough, the scientists were able to pick out four useful pieces — modules made up of around 60 to 100 amino acids. Each one can carry electrons or carry out reactions but uses different metals (such as copper, iron, nickel or manganese) or puts those metals in different configurations.
The scientists think the oldest of the four is the one with a cube-shaped cluster of four irons and four sulfurs. That’s because this subunit would be very handy for harvesting energy around hydrothermal vents, which are known to host life-like chemical reactions even when living things aren’t around. That kind of chemistry could have made it much easier for microbes with the right kinds of electron-shuttling proteins to thrive, the thinking goes.
These ideas will have to be tested further in the lab, Nanda said. In the meantime, the scientists expect there are more Lego-like subunits beyond the four they’ve identified.
“We really are only looking at a very small subset of proteins, the ones that are involved in electron transfer,” Nanda said.
These smash-and-search methods could be used to find shared building blocks within other groups of proteins beyond the electron-transfer group, he added.
The scientists also want to chop up those four subunits of 60 to 100 amino acids and find even smaller, simpler essential pieces within them, he said. The simpler the unit, the closer it might be to those primordial proteins.
Beyond helping scientists understand the proteins from life’s distant past, this research could one day help scientists develop proteins of the future.
Proteins are already being designed for industrial and therapeutic uses, but this new technology could improve the process, scientists said. It could even be used to design proteins that split water — which could lead to a cheap way of generating clean-burning hydrogen fuel.
And as computers continue to shrink, specially designed electron-transfer proteins could potentially help engineers build smaller transistors and diodes and capacitors — perhaps even find a way to make them self-assemble into circuits, Nanda said.
“Then you could start to make electronic circuits not by lithography and printing these things onto chips,” he said, “but maybe have bacteria synthesizing small electrical circuits for you and doing these sorts of things in a sustainable way.”
Follow @aminawrite on Twitter for more science news and “like” Los Angeles Times Science & Health on Facebook.
MORE IN SCIENCE
Forget concussions. The real risk of CTE comes from repeated hits to the head, study shows
2017 was one of the three hottest years on record, NASA and NOAA scientists say
This new blood test can detect early signs of 8 kinds of cancer